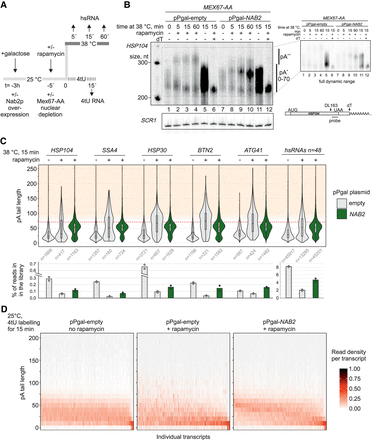
Nab2p controls nuclear mRNA pA tail length. (A) Experimental scheme to measure nuclear mRNA pA tail lengths. mRNA export was conditionally blocked in MEX67-AA cells, overexpressing Nab2p or not, 5 min before transferring cells to 38°C (dark line on top) or initiating 4tU labeling (dashed dark line). RNA samples were collected at the indicated times after heat treatment or initiation of 4tU labeling. (B) RNase H/Northern analysis of HSP104 RNA 3′ ends from MEX67-AA cells subjected to different 38°C incubation periods, conditionally depleted of nuclear Mex67p (rapamycin −/+) and expressing endogenous levels of Nab2p (“pPgal-empty”) or overexpressing Nab2p from a plasmid under control of a galactose inducible promoter (“pPgal-NAB2”) as indicated. (Bottom right panel) HSP104 RNA was detected using an oligonucleotide probe close to the pA site and, to increase resolution, transcripts were internally cleaved with RNase H targeted by the DL163 DNA antisense oligo, annealing 230 nt upstream of the pA site. A subset of RNase H treatments further included a dT18 oligo (dT −/+) to trim away pA tails. SCR1 RNA was probed as a loading control. Migration of HSP104 RNA harboring normal (pA+, 0–70 As) and hyperadenylated (pA++, >70 As) pA tails are indicated. Pixel intensities have been linearly enhanced to emphasize the hyperadenylated RNA. The top right panel inset shows the same exposure in full pixel intensity range. (C) Quantification of hsRNA pA tail lengths by direct RNA sequencing. Polyadenylated RNA was extracted from MEX67-AA cells shifted for 15 min to 38°C without (“−”) or with (“+”) a 5-min pretreatment with rapamycin, and without (gray shaded violins) or with (green violins) ectopic Nab2p expression as in B. pA tail lengths are shown for five individual hsRNAs and a meta-analysis of the top 48 most heat-induced mRNAs. pA tails >70 As (orange-shaded area above the red dashed line) were defined as hyperadenylated. The number of sequence reads in each group is denoted below the violins. Bar plots below the graphs display the mean percentage of reads from shown transcripts within the sequenced DRS libraries. Individual values from two independent replicates are shown as dots. (D) pA tail length distributions of 4tU RNAs. Reads from each RNA were grouped based on their pA tail lengths in 10-A bins, and bin values were then normalized to the total number of reads for each transcript. The resulting fraction of reads in each bin is represented using the indicated color gradient. Only transcripts containing >20 reads in the “pPgal-NAB2 + rapamycin” sample were included in the analysis (n = 821). Transcripts were sorted by their median pA tail length in the right panel.












