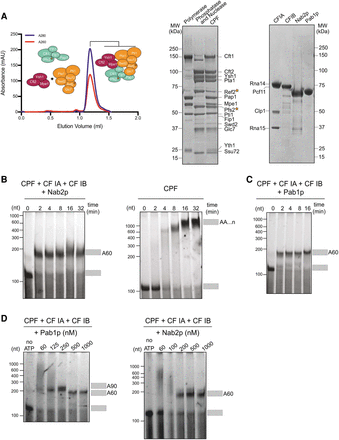
Nab2p and Pab1p control pA tail length in a reconstituted polyadenylation system. (A, left) Size exclusion chromatogram of recombinant CPF reconstitution from the polymerase module (mint) and combined phosphatase (orange) and nuclease (magenta) modules. Tagged proteins are indicated with a star. (Right) Coomassie-stained SDS-PAGE analysis of purified proteins and complexes used for the polyadenylation assays. (B–D) Denaturing RNA gels of polyadenylation assays using a fluorescently labeled 120-nt pcCYC1 RNA substrate (final concentration 100 nM). Adenylated and nonadenylated forms of the substrate are indicated as gray rectangles. All assays were repeated at least three times, and representative gels are shown. (B) Time course analysis of polyadenylation by CPF in the presence or absence of Nab2p (600 nM) and cleavage factors CF IA and CF IB, as indicated above the gel panels. (C) Time-course analysis of polyadenylation by CPF in the presence of Pab1p (1 µM) and the cleavage factors CF IA and CF IB. (D) Effect of Pab1p (left panel) and Nab2p (right panel) concentrations on CPF/CF IA/CF IB-synthesized pA tail lengths. Polyadenylation reactions were initiated by the addition of ATP and stopped after 8 min. First gel lanes at the left show reactions in the absence of ATP. Time courses for corresponding reactions are displayed in Supplemental Figure S3, A and D.












