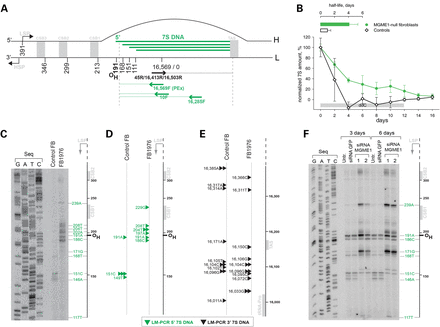
Loss of MGME1 modifies the 5′ ends of DNA species in the NCR. (A) The human mitochondrial NCR. The position of the main 5′ ends of 7S DNA found in this and other studies (6–8) are indicated. HSP and LSP—heavy- and light-strand promoter, respectively. CSB—conserved sequence block, TAS—termination associated sequences. Positions of primers used in the PEx and LM-PCR analysis are given. (B) 7S DNA levels in control and patient fibroblasts quantified by qPCR during ddC treatment for the indicated time (grey bar). Two controls and two patient samples were investigated, each determined in three independent reactions. Half-life times were determined by nonlinear regression analysis of qPCR data assuming simple exponential decay kinetics (inset). (C) Primer extension mapping of DNA 5′ ends in human control and patient fibroblasts (FB1976). A radioactively labelled primer annealing at nt position 16 569–18 of human mtDNA was extended using total DNA preparations treated with RNase A during cell lysis. (D) DNA 5′ ends detected by LM-PCR in controls and FB1976. Further details are provided in Supplementary Material, Figure S2. (E) DNA 3′ ends in the vicinity of the TAS sequence detected by LM-PCR in human control and patient fibroblast (FB1976). (F) PEx mapping of DNA 5′ ends in the human mtDNA NCR of HeLa cells transfected with siRNA to MGME1 or siRNA to GFP for 3 or 6 days. The experiment was performed as per (C). ‘Untr’—non-transfected control.



