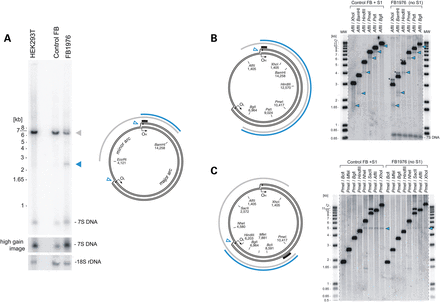
Loss of MGME1 results in accumulation of an 11-kb sub-genomic mtDNA fragment. (A) (left) Total DNA from HEK293T cells, control and patient fibroblasts (FB1976, cell passage 8) was digested with BamHI and EcoRI and analysed by 1D Southern blot with a probe specific for the NCR (mtDNA region: 16 341–151). 18S rDNA was used as a loading control. Blue arrowhead—2.5-kb sub-1n fragment observed recurrently for FB1976. (right) Schematic representation of the source of the sub-genomic fragment. Grey arc—the full-length genomic fragment. Empty blue arrowheads—chromosomal breakage of replication intermediates near OH and OL. Blue arc—2.5-kb sub-1n fragment. Black bar—the probe. (B) (left) The restriction map of human mtDNA indicating sites relevant for the analysis. Blue arc—the sub-genomic fragment near OH. Black bar—the probe (mtDNA region: 16 341–151). (right) Total DNA from control fibroblasts treated with S1 nuclease or from FB1976 was analysed by 1D Southern blotting using indicated restrictases. Blue arrowheads—the sub-genomic fragments generated by S1 nuclease treatment of control cells or present in FB1976. Asterisks indicate the main genomic restriction fragment with residual 7S DNA still bound (Supplementary Material, Fig. S5B). (C) (left) The restriction mapping of the sub-genomic fragments near OL (blue arc). Black bar—the probe (mtDNA region: 9 610–10 218). (right) 1D Southern blot analysis as per (B) using restriction enzymes as indicated. The remaining indications as per (B).



