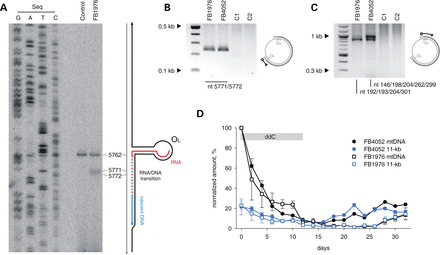
Blunt ends and prolonged persistence of the 11-kb sub-genomic mtDNA fragment in MGME1-deficient fibroblasts. (A) PEx mapping of DNA 5′ ends near OL in control and FB1976. A radioactively labelled primer annealing at nt position 5915–5898 of human mtDNA was extended using total DNA preparations treated with RNase A. Right: the predicted hairpin structure at OL and the current model of OL-specific initiation of lagging-strand mtDNA synthesis (30). (B) Double-stranded blunt ends close to OL in patient fibroblasts detected by LM-PCR. Amplification was performed using a linker-specific and an mtDNA-specific primer. The ends were identified by sequencing 12 single-molecule amplicons. C1, fibroblasts from an 11-year-old healthy control; C2, fibroblasts from a 39-year-old healthy control. (C) Double-stranded blunt ends in the vicinity of OH. Note that while 7S DNA 5′ ends are present in this region both in patients and in controls (Fig. 1F, Supplementary Material, Fig. S2), blunt dsDNA ends are specific for MGME1-deficient patients. Positions of mapped ends are indicated. (D) The presence of the 11-kb sub-genomic mtDNA fragment in MGME1-deficient fibroblasts mtDNA during depletion-repopulation experiment. Fibroblasts from patients were treated with ddC for 12 days (indicated by grey bar) followed by culturing without the inhibitor for 20 days. Genomic (black) and sub-genomic (blue) fragment amounts were quantified by qPCR and normalized to copy numbers of complete ds mtDNA molecules measured at the beginning of the ddC treatment. Error bars—SD values. n = 3.



