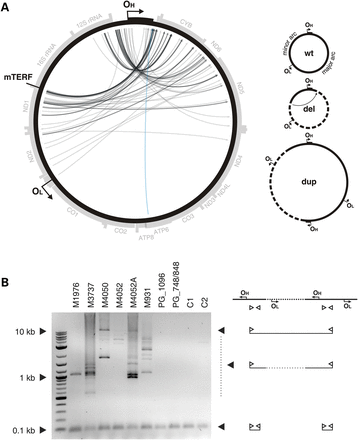
Unusual mtDNA rearrangements in MGME1 patients. (A) Circos representation of detected mtDNA breakpoints in patients with pathogenic MGME1 mutations. Arrowheads indicate the orientation of the deletions. The part of the genome that is deleted spans counter clockwise from the root to the head of the arrow (as indicated by dashed line in the panel ‘del’ of the scheme on the right). A detailed list of the detected breakpoints is available in Supplementary Material, Table S2. Breakpoints that conform to regular major arc deletions are marked in blue; grey arcs indicate breakpoints that remove OL. Possible interpretations of the breakpoints are shown in the scheme on the right. del, deleted mtDNA molecules; dup, partially duplicated mtDNA; wt, wild-type. (B) Partially duplicated mtDNA molecules in skeletal muscle of MGME1 patients as detected by long-extension PCR. The scheme on the right indicates the interpretation of the detected bands. Bands at 0.1 kb represent short PCR fragments deriving from primers annealing at neighbouring positions. Bands at 0.5 kb and above originate from partial or complete duplications of mtDNA. PG_1096, 6-year-old patient with homozygous mutation R1096C in POLG; PG_748, 12-year-old patient with compound heterozygous mutations W748S and G848S in POLG; C1, 35-year-old healthy control; C2, 63-year-old healthy control.



