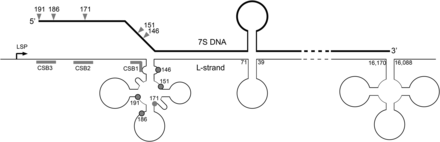
Hypothetical secondary structures in the D-loop region and a role for MGME1. Secondary structures are depicted as forming in the mtDNA NCR region, according to the predictions of Brown et al. (41). Formation of a cloverleaf structure in the mtDNA L-strand in the OH region could prevent the 5′ end of 7S DNA from annealing to the template strand, leaving 7S DNA displaced as along 5′ ssDNA flap. The region covered by this cloverleaf structure encompasses almost all of the major sites of 7S DNA 5′ ends. If MGME1 is directly involved in the processing of 7S DNA ends, then the 5′ end of 7S DNA must be displaced into an ssDNA flap in order to be a good substrate for MGME1, according to the in vitro characterization presented here. Displacement of the 5′ end of 7S DNA by a secondary structure as shown above could serve this purpose.



