Phosphorylation of spore coat proteins by a family of atypical protein kinases
- Kim B. Nguyena,1,
- Anju Sreelathab,1,
- Eric S. Durranta,
- Javier Lopez-Garridoc,
- Anna Muszewskad,
- Małgorzata Dudkiewicze,
- Marcin Grynbergf,
- Samantha Yeeb,
- Kit Poglianoc,
- Diana R. Tomchickg,
- Krzysztof Pawłowskie,
- Jack E. Dixona,h,i,2, and
- Vincent S. Tagliabraccib,2
- aDepartment of Pharmacology, University of California, San Diego, La Jolla, CA 92093;
- bDepartment of Molecular Biology, University of Texas Southwestern Medical Center, Dallas, TX 75390;
- cDivision of Biological Sciences, University of California, San Diego, La Jolla, CA 92093;
- dDepartment of Microbial Biochemistry, Institute of Biochemistry and Biophysics, Polish Academy of Sciences, 02106 Warsaw, Poland;
- eDepartment of Experimental Design and Bioinformatics, Warsaw University of Life Sciences, 02787 Warsaw, Poland;
- fDepartment of Biophysics, Institute of Biochemistry and Biophysics, Polish Academy of Sciences, 02106 Warsaw, Poland;
- gDepartments of Biophysics and Biochemistry, University of Texas Southwestern Medical Center, Dallas, TX 75390;
- hDepartment of Cellular and Molecular Medicine, University of California, San Diego, La Jolla, CA 92093;
- iDepartment of Chemistry and Biochemistry, University of California, San Diego, La Jolla, CA 92093
-
Contributed by Jack E. Dixon, April 13, 2016 (sent for review March 23, 2016; reviewed by John Scott and Andrey Shaw)
Significance
The posttranslational modification of proteins with a molecule of phosphate, termed protein phosphorylation, is a mechanism used by cells to regulate cellular activities. Protein phosphorylation occurs in all life forms and is catalyzed by a superfamily of enzymes known as protein kinases. Using bioinformatics, we have identified a family of spore coat protein (Cot) kinases, which are related to the secreted kinase, family with sequence similarity 20C (Fam20C). The founding member of this family is CotH from the spore-forming bacterium Bacillus subtilis. We show that CotH-dependent phosphorylation of the spore proteins CotB and CotG is necessary for the proper germination of spores. Because several CotH-containing organisms are human pathogens, our work has important clinical implications to combat human diseases, such as anthrax.
Abstract
The modification of proteins by phosphorylation occurs in all life forms and is catalyzed by a large superfamily of enzymes known as protein kinases. We recently discovered a family of secretory pathway kinases that phosphorylate extracellular proteins. One member, family with sequence similarity 20C (Fam20C), is the physiological Golgi casein kinase. While examining distantly related protein sequences, we observed low levels of identity between the spore coat protein H (CotH), and the Fam20C-related secretory pathway kinases. CotH is a component of the spore in many bacterial and eukaryotic species, and is required for efficient germination of spores in Bacillus subtilis; however, the mechanism by which CotH affects germination is unclear. Here, we show that CotH is a protein kinase. The crystal structure of CotH reveals an atypical protein kinase-like fold with a unique mode of ATP binding. Examination of the genes neighboring cotH in B. subtilis led us to identify two spore coat proteins, CotB and CotG, as CotH substrates. Furthermore, we show that CotH-dependent phosphorylation of CotB and CotG is required for the efficient germination of B. subtilis spores. Collectively, our results define a family of atypical protein kinases and reveal an unexpected role for protein phosphorylation in spore biology.
Protein phosphorylation is a nearly universal mode of cellular regulation and is catalyzed by a large superfamily of enzymes known as protein kinases (1). In eukaryotes, most protein kinases transfer a molecule of phosphate from ATP onto Ser, Thr, and Tyr residues. In contrast, bacterial kinases frequently modify His residues on proteins as part of a two-component system (2). Nevertheless, prokaryotic kinases, such as the bacterial persistence kinase, high-persistence factor A (HipA), and the proinflammatory kinase, cell translocating kinase A (CtkA), can modify Ser and Thr residues on proteins as well (3, 4).
We recently identified a small family of eukaryotic protein kinases that act on their substrates within the lumen of the secretory pathway (5, 6). These enzymes share limited sequence similarity with canonical protein kinases, and some of them phosphorylate proteins, whereas others phosphorylate proteoglycan substrates (7⇓–9). One member, family with sequence similarity 20C (Fam20C), is the physiological Golgi casein kinase (5, 10). Fam20C phosphorylates hundreds of secreted proteins with a marked preference for Ser residues within the consensus sequence Ser-x-Glu/pSer (11⇓–13). Human mutations in FAM20C cause Raine syndrome, an often-fatal osteosclerotic bone disorder (14). Fam20A, a paralog of Fam20C, is a pseudokinase that binds Fam20C and stimulates its kinase activity (15). Consequently, loss-of-function mutations in FAM20A cause enamel renal syndrome and amelogenesis imperfecta in humans (16, 17). Fam20B is a proteoglycan kinase that phosphorylates the 2-hydroxyl of a xylose residue within the tetrasaccharide linkage region of proteoglycans (7). Fam20B-dependent xylose phosphorylation stimulates glycosaminoglycan assembly by enhancing the activity of galactosyl transferase II, an enzyme required for the biosynthesis of proteoglycans (18).
We previously noted sequence and structural similarities between HipA/CtkA and the eukaryotic Fam20C-like secretory pathway kinases described above (5, 19). While examining sequences related to HipA, we noticed limited but significant sequence similarity between HipA and a family of proteins known as spore coat protein H (CotH). CotH orthologs are found in many bacterial and eukaryotic species, including several human pathogens, such as the spore-forming bacterium Bacillus anthracis and the spore-forming fungi Rhizopus oryzae, the causative agents of anthrax and mucormycosis, respectively (20, 21). CotH is a component of the spore coat, a multilayered shell consisting of ∼70 proteins that are assembled into layers by regulatory proteins referred to as morphogenetic factors (22). CotH is one such morphogenetic factor and is involved in the assembly of at least nine other spore coat proteins in the model spore-forming organism Bacillus subtilis (23⇓–25). CotH appears to maintain the integrity of the spore coat because loss of CotH in B. subtilis is detrimental to the germination efficiency of the spores (24⇓–26). However, the molecular mechanisms through which this occurs are unclear.
Here, we show that B. subtilis and Bacillus cereus CotH are protein kinases. Examination of the genes neighboring cotH in B. subtilis led us to identify two spore coat proteins, CotB and CotG, as CotH substrates. Moreover, CotH kinase activity is required for the efficient germination of spores in B. subtilis. We have solved crystal structures of the apo- and nucleotide-bound CotH from B. cereus, which reveal an atypical protein kinase-like (PKL) fold and highlight unique residues involved in nucleotide binding and catalysis. Collectively, our results uncover a family of atypical spore coat protein kinases and reveal a previously unappreciated role for protein phosphorylation in spore biology.
Results
CotH Exhibits Significant Sequence Similarity to Protein Kinases.
Examination of proteins with distant sequence similarities to the Fam20C secretory kinases led us to hypothesize that the CotH family might function as atypical protein kinases (Fig. 1A and Figs. S1 and S2). Structural prediction analyses suggest low but significant similarity between CotH proteins and authentic kinases (Materials and Methods). Galperin et al. (27) have briefly noted this similarity as well.
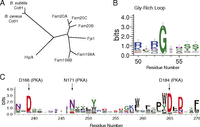
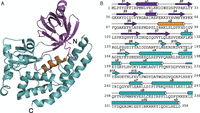
CotH proteins share sequence similarity with known protein kinases. (A) Phylogenetic tree depicting the eukaryotic secretory pathway kinases and related prokaryotic proteins: Fam20A/B, Fam198A/B, four-jointed box 1 (Fjx1), and HipA. Sequence logos were created by WebLogo, using MUSCLE multiple sequence alignment of 165 CotH homologs depicting the loop between strands β1 and β2 (B, Gly-rich loop) and the active site residues (C).
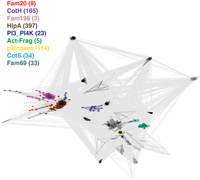
CotH proteins share sequence similarity with protein kinases. CLANS clustering analysis is represented graphically as the network of BLAST-derived sequence similarities (edges) between representatives of known kinase families (dots). All PKL clan families are included, including those clan families predicted to have a PKL fold, such as the Fam69 family. Magenta, CotH; red, Fam20; pink, Fam198; brown, HipA; dark blue, PI3_PI4K; light green, actin-fragmin kinases; light blue, CotS; cyan, Fam69; yellow, pkinase (typical eukaryotic Ser/Thr and Tyr kinases).
Multiple sequence alignment of selected CotH proteins and other kinases. The alignment was generated with PROMALS3D (77) and includes selected CotH homologs (representatives of clusters seen in Fig. 7), with additional related kinases [Fam20C, phosphatidylinositol 4-kinase IIα (PI4K), HipA, CtkA, PKA, and CotS]. National Center for Biotechnology Information GeneInfo identifier (gi) numbers are shown.
A sequence logo built using 165 representative CotH homolog sequences illustrates the conservation of some features typical of kinases, including a conserved RG motif in the β1-β2 ATP-binding loop (Gly-rich loop) (Fig. 1B) and conserved active site residues corresponding to D166, N171, and D184 of the prototypical protein kinase, PKA (28) (Fig. 1C). In contrast, some features typically found in canonical kinases are absent. For example, there are no clear equivalents of K72 and E91, which form an ion pair in PKA, and a conserved PWD[LF]D motif replaces the metal-binding DFG motif in the active site region.
B. subtilis and B. cereus CotH Are Active Protein Kinases.
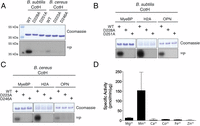
B. subtilis and B. cereus CotH are ∼40-kDa proteins that share about 60% sequence similarity. To determine whether the CotH proteins are active kinases, we expressed B. subtilis and B. cereus CotH in Escherichia coli as 6X-His-Sumo–fusion proteins and purified the proteins by nickel-nitrilotriacetic acid (Ni-NTA) affinity chromatography. We also purified recombinant CotH proteins that harbor mutations within predicted active site catalytic residues, including B. subtilis D228A and B. cereus D223A (predicted PKA D166 equivalent) and B. subtilis D251A and B. cereus D246A (predicted PKA D184 equivalent). Recombinant B. subtilis and B. cereus CotH, but not the inactive mutant proteins, underwent autophosphorylation in the presence of γ-32P[ATP] (Fig. 2A). Moreover, both B. subtilis and B. cereus CotH, but not the mutants, phosphorylated the generic protein kinase substrates myelin basic protein (MyeBP), histone H2A (H2A), and osteopontin (OPN) (Fig. 2 B and C). Most protein kinases require a divalent cation to orient the nucleotide phosphates within the active site for catalysis (29). B. subtilis CotH displays a preference for Mn2+ as the activating divalent cation (Fig. 2D). Collectively, these results demonstrate that CotH proteins are protein kinases.
B. subtilis and B. cereus CotH are active protein kinases. (A) Incorporation of 32P from [γ-32P]ATP into B. subtilis and B. cereus wild-type (WT) CotH or the indicated mutants. Reaction products were separated by SDS/PAGE and stained with Coomassie blue. The incorporated radioactivity was visualized by autoradiography. Incorporation of 32P from [γ-32P]ATP into MyeBP, H2A, and OPN by B. subtilis wild-type CotH or the indicated mutants (B) and B. cereus wild-type CotH or the indicated mutants (C). Reaction products were analyzed as in A. (D) Incorporation of 32P from [γ-32P]ATP into a peptide substrate, CotG(88–100), which is derived from the CotH substrate CotG (described in Fig. 6) by B. subtilis CotH. The reactions were carried out in the presence of 5 mM MgCl2, MnCl2, CaCl2, CoCl2, FeCl2, or ZnCl2, and the reaction products were spotted on P81 phosphocellulose filter papers and terminated by immersion in H3PO4. Filter papers were washed, and incorporated radioactivity was quantified by scintillation counting.
The Crystal Structure of B. cereus CotH Reveals an Atypical PKL Fold with a Unique Mode of Nucleotide Binding.
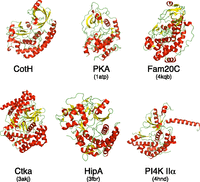
We solved the crystal structure of full-length B. cereus CotH at a resolution of 1.6 Å (Fig. 3A and Table S1). B. cereus CotH has an atypical PKL fold containing 11 α-helices, 3 310 helices, and 13 β-strands (Fig. 3B). Common among all protein kinases is the presence of two structurally and functionally distinct lobes, termed the N- and C-lobes (30). The N-lobe in B. cereus CotH contains three helices and seven β-sheets, the latter of which make up a β-barrel–like subdomain. The larger C-lobe consists of α-helices and a distinctive four-stranded β-sheet. Three relatively large helices (α8–α10 ) cover one side of the molecule and make multiple contacts with the N-lobe. Database searches using the Dali server (31) indicate that B. cereus CotH is most similar to HipA and the phosphatidylinositol 4-kinase family (Fig. S3).
Crystal structure of B. cereus CotH reveals an atypical PKL fold. (A) Ribbon representation of B. cereus CotH. The N- and C-lobes are shown in magenta and teal, respectively. The α2 helix (PKA αC equivalent) is highlighted in orange. (B) Amino acid sequence of B. cereus CotH depicting the secondary structural elements, color-coded as in A. The 310 helices (η) are also shown.
Data collection and refinement statistics of CotH structures
Structural comparison of B. cereus CotH with PKA and selected atypical kinases. The structures of B. cereus CotH, PKA (PDB ID code 1ATP), Caenorhabditis elegans Fam20C (PDB ID code 4KQB), CtkA (PDB ID code 3AKJ), HipA (PDB ID code 3FBR), and PI4K IIα (PDB ID code 4HND) are shown. The α-helices are colored in red, and the β-strands are colored in yellow.
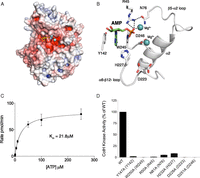
We also attempted to crystallize B. cereus CotH in complex with Mg2+/ADP, but the resulting electron density map at a resolution of 1.4 Å is consistent with Mg2+/AMP in the active site (Fig. 4A). The protein backbone of the Mg2+/AMP structure is virtually identical to the apo structure, with only a few residues displaying significant movement upon nucleotide binding. As in all kinases, the nucleotide sits in a cleft between the N-lobe and C-lobe (Fig. 4A). In most protein kinases, the adenine moiety of ATP is nestled within a pocket of hydrophobic residues (32). However, in B. cereus CotH, the adenine moiety of AMP is sandwiched between two aromatic residues, Tyr142 from β8 and Trp245 from the β13-α7 loop (Fig. 4B). Both B. subtilis and B. cereus CotH displayed a Km for ATP of ∼20 μM (Fig. 4C), a value that falls well within the range of most Ser/Thr kinases. As expected, mutation of the corresponding residues in B. subtilis CotH, Tyr147 and Trp250, to Ala completely abolished kinase activity (Fig. 4D). These results support a critical and unique role for these residues in ATP binding and catalysis by CotH family kinases.
Mg2+/AMP-bound structure of B. cereus CotH reveals a unique mode of nucleotide binding and highlights residues involved in catalysis. (A) Surface representations illustrating the electrostatic potential of B. cereus CotH depicting AMP (yellow sticks) and Mg2+ (cyan spheres) bound in a cleft between the N- and C-lobes of the kinase. The electrostatic potentials were calculated with the APBS2.1 plug-in of PyMOL (65). The gradients of electrostatic potentials shown ranged from ≥−5 kbT/ec (red) to ≤+5 kbT/ec (blue), where kb is Boltzmann’s constant, T is temperature in degrees Kelvin, and ec is the charge of an electron. (B) Expanded image of the nucleotide-binding pocket showing the detailed molecular interactions important for nucleotide binding and catalysis. The AMP molecule and the two Mg2+ ions are shown as green sticks and cyan spheres, respectively. The salt bridge and hydrogen bond interactions are shown as dashed lines. (C) Kinetic analysis depicting the concentration dependence of Mn2+/ATP on the rate of phosphate incorporation into CotG(88–100) by B. subtilis CotH. (Inset) Km for Mn2+/ATP is indicated. Reaction products were analyzed as in Fig. 2D. (D) Kinase activity of B. subtilis CotH and active site mutants was analyzed as in Fig. 2D. Activity is expressed relative to activity of the wild-type enzyme. The amino acids in brackets indicate the corresponding residues in B. cereus CotH.
The N-lobe in B. cereus CotH is quite different from what is commonly observed in canonical protein kinases. Most notably, the N-lobe contains a seven-stranded β-barrel–like subdomain filled with hydrophobic residues. The α2 helix (equivalent to αC in PKA) lies between β5 and β6 and extends into the C-lobe. The αC helix is present within the N-lobe in virtually all protein kinases and plays an essential role in regulating kinase activity by dynamically rearranging the active site upon nucleotide binding (33). In B. cereus CotH, α2 occupies a stable position in the center of the molecule in both the apo- and Mg2+/AMP-bound structures and is unlikely to play a regulatory role.
A short loop is present in B. cereus CotH that lies between β3 and η1, and is analogous to the Gly-rich loop in canonical protein kinases. A conserved Arg (Arg45) extends into the active site and coordinates the α-phosphate of AMP. Virtually all protein kinases contain a highly conserved Lys in β3 and a Glu in αC that form a salt bridge (Lys72 and Glu91 in PKA) (28). This interaction is considered a hallmark of an activated protein kinase because Lys72 coordinates the α- and β-phosphates of ATP and Glu91 functions to stabilize the conformation of the Lys (33). In B. cereus CotH, Arg45 is stabilized by a hydrogen bond from Asn76 in β5, and this interaction functionally appears to replace the salt bridge of canonical kinases. Consistently, mutation of the corresponding residues in B. subtilis CotH, Arg50, and Asn81 to Ala severely reduced activity of the kinase (Fig. 4D).
The C-lobe in B. cereus CotH is also very different from most protein kinases and harbors the majority of the residues involved in catalysis. In canonical kinases, a conserved “DFG” motif acts to bind divalent cations to orient the nucleotide phosphates (29). A highly conserved (W/Y/F)DxD motif is present in the CotH family and appears to play a similar role. Asp246 (Asp184 in PKA) in B. cereus CotH lies within a large loop that connects β13 to α7 and binds an Mg2+ ion (Fig. 4B). The large aromatic residue within this motif (Trp245 in B. cereus CotH), together with Tyr143, orients the adenine ring as described above. Mutation of the analogous residue in B. subtilis CotH, Asp251 to Ala, eliminated activity of the kinase (Fig. 4D).
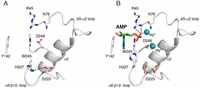
The catalytic loop is positioned between α6 and β12 and contains the putative catalytic Asp223 (Asp166 in PKA). Mutation of this Asp (Asp228 in B. subtilis CotH) to Ala eradicates kinase activity (Fig. 4D). In the apo-structure, Asp223 forms an ion pair with His227 and this interaction is disrupted upon nucleotide binding (Fig. S4). In the AMP-bound structure, His227 moves toward the nucleotide-binding pocket and may interact with the phosphates of ATP. Mutation of His227 (His232 in B. subtilis CotH) to Ala significantly reduces activity, thus underscoring the importance of this residue in catalysis (Fig. 4D). Collectively, the crystal structure of B. cereus CotH reveals a unique mechanism by which atypical protein kinases can bind ATP and catalyze phosphotransfer. These results further highlight the structural diversity of the protein kinase superfamily.
Comparison of the active sites in the apo- and AMP/Mg2+ structures. Expanded images of the nucleotide-binding pocket in the apo structure (A) and the AMP/Mg2+ structure (B) are shown. The AMP molecule and the two Mg2+ ions are shown as green sticks and cyan spheres, respectively. Interactions are shown as dashed lines.
CotH Kinase Activity Is Required for Efficient Germination of B. subtilis Spores.
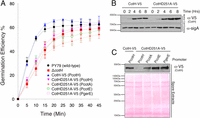
In B. subtilis, CotH is a structural component of the spore coat and is required for the development of spores that are able to germinate efficiently (25). We generated a cotH-null strain of B. subtilis (ΔcotH), induced spore formation by nutrient deprivation, and purified the resulting spores to perform germination assays (a list of strains is provided in Table S2; see also Table S3). As expected, cotH-null spores germinated markedly less efficiently than wild-type spores when stimulated with the germinant L-alanine (Fig. 5A). To establish the importance of CotH kinase activity for germination efficiency, we reintroduced V5-tagged wild-type CotH or the D251A inactive mutant at the amyE locus and monitored germination efficiency (34). Ectopic expression of wild-type CotH, but not the D251A mutant, rescued the germination defect in the cotH-null strain (Fig. 5A).
CotH kinase activity is required for the proper germination of B. subtilis spores. (A) Germination efficiency was analyzed in spores derived from the strains displayed in Table S2. Germination was induced by L-alanine and measured as the percentage of loss of optical density at 580 nm. (B) Protein immunoblotting of mother cell extracts depicting the time-dependent expression of V5-tagged wild-type CotH and the D251A mutant following induction of sporulation. α-SigA immunoblots are shown as a loading control. (C, Upper) Protein immunoblotting of spore coat extracts depicting the incorporation of V5-tagged CotH into the spore coat. (C, Lower) Ponceau S-stained membrane is shown as a loading control.
B. subtilis strains used in this study
List of primers used in this study
We then monitored CotH protein levels in extracts from sporulating cultures at different time points after sporulation induction. We detected expression of both the wild-type and D251A mutant proteins about 2 h postinduction (Fig. 5B). By 6-h postinduction, the protein level of the D251A mutant was slightly elevated relative to the wild-type protein. To determine if both wild-type CotH and the D251A mutant were efficiently incorporated to the coat, we isolated coat proteins from purified spores and performed V5-immunoblotting. Wild-type CotH, but not the D251A mutant, was efficiently incorporated into the spore coat (Fig. 5C). Therefore, to determine whether the germination defect in cotH-null spores was due to loss of kinase activity, reduced incorporation of CotH into the coat, or a combination of both, we overproduced CotH D251A by placing the coding sequence at the amyE locus under the control of the strong promoters of CotA, CotE, or GerE (Table S2). Protein immunoblotting of spore coat extracts revealed that expression of the catalytically inactive CotH from each of these promoters was sufficient to incorporate the mutant protein into the spore coat (Fig. 5C). Similar to the cotH-null strain, spores purified from the CotH D251A-overproducing strains germinated more slowly than spores expressing wild-type CotH (Fig. 5A). Thus, loss of CotH kinase activity, rather than aberrant assembly of CotH into the spore, is the likely cause for impaired germination of B. subtilis spores.
B. subtilis CotH Regulates Spore Germination by Phosphorylation of the Spore Coat Proteins CotB and CotG.
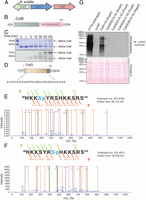
The cotH gene clusters with cotB and cotG in B. subtilis, and all three genes encode proteins that reside in the proteinaceous coat of the spore (35) (Fig. 6A). The cotB encodes a predicted 43-kDa polypeptide that migrates in SDS/PAGE with an apparent molecular mass of 66 kDa in sporulating bacteria (36). Interestingly, CotB migrates as a 43-kDa species in cotH-null spores, suggesting that CotH promotes the formation of the more slowly migrating 66-kDa form of CotB (26, 36). CotB has a C-terminal domain that is enriched in Ser, Lys, and Arg residues (Fig. 6B). We hypothesized that the CotB modification observed in vivo was due to phosphorylation of CotB by CotH, thus accounting for the apparent 23-kDa decrease in molecular mass of CotB in cotH-null spores (26, 36). We purified full-length recombinant CotB from E. coli and performed in vitro kinase assays. CotH phosphorylated CotB in a time-dependent manner and was able to convert the 43-kDa species of CotB to the more slowly migrating 66-kDa species as assessed by gel electrophoresis (Fig. 6C). We conclude that the 66-kDa form of CotB in sporulating bacteria is generated by multisite phosphorylation catalyzed by CotH.
B. subtilis CotH phosphorylates the spore coat proteins CotB and CotG. (A) B. subtilis cotH gene forms a cluster with two genes that encode spore coat proteins cotB and cotG. (B) Schematic representation of B. subtilis CotB depicting the Ser/Arg/Lys-rich C-terminal region. (C) Time-dependent incorporation of 32P from [γ-32P]ATP into 6× His-tagged CotB by B. subtilis CotH. Reaction products were separated by SDS/PAGE and visualized by Coomassie staining (Upper), and radioactivity was detected by autoradiography (Lower). (D) Schematic representation of B. subtilis CotG depicting the nine tandem repeats in the protein. Representative tandem mass spectroscopy (MS/MS) fragmentation spectra depicting Ser4 (E) and Ser7 (F) phosphorylation of CotG(88–100) by CotH is shown. (G, Upper) Protein immunoblotting of B. subtilis spore coat extracts using a phosphospecific PKC substrate antibody depicting the phosphorylation of spore coat proteins in the different strains. (G, Lower) Ponceau S-stained membrane is shown as a loading control. The asterisks depict proteins that we infer are CotB and CotG based on their molecular masses.
CotH appears to stabilize CotG because strains lacking CotH fail to incorporate CotG into the spore coat (36). However, the mechanism by which CotH stabilizes CotG is unknown. CotG encodes an ∼24-kDa protein consisting of nine tandem repeats of the amino acid sequence: (H/Y)-K-K-S-(H/Y/F/C)-(R/C)-(T/S)-(H/Y)-K-K-S-R-S (Fig. 6D). The protein is rich in Ser residues (39 Ser) and is highly basic, because nearly half of its residues are Arg, Lys, or His. A recent proteomics experiment has shown that B. subtilis CotG is phosphorylated on multiple residues within the tandem repeats (26). In fact, the CotG ortholog in B. anthracis, ExsB, has been described as the most extensively phosphorylated bacterial protein characterized to date (37).
To test CotG as a potential substrate for CotH, we attempted to produce full-length recombinant CotG in E. coli; however, despite several attempts, we were unable to purify the protein successfully. Therefore, we synthesized a B. subtilis CotG peptide corresponding to a natural sequence occurring in one of the repeats: 88HKKSYRSHKKSRS100, hereafter referred to as CotG(88–100). As shown in Figs. 2D and 4 C and D, B. subtilis CotH phosphorylated CotG(88–100). Mass spectrometry analysis of phosphorylated CotG(88–100) revealed that peptides contained phosphate on either Ser4 or Ser7 (Fig. 6 E and F). However, we did not detect any peptides in which both Ser4 and Ser7 were simultaneously phosphorylated. Consistent with these results, using γ-32P[ATP] of defined specific activity and quantifying the incorporated 32P, we calculated a stoichiometry of ∼1 mol of phosphate incorporated per mole of CotG(88–100) following exhaustive phosphorylation by B. subtilis CotH.
Both CotB and CotG are phosphorylated on Ser residues adjacent to basic amino acids. In fact, 37 of the 39 Ser residues in CotG and more than half of the 57 Ser residues within the C terminus of CotB contain a Lys or Arg at the n − 2 or n + 2 position (R/K-x-S-x or x-S-x-R/K). This motif resembles the consensus recognition sequence for the eukaryotic protein kinase C (PKC). We therefore purified spores from wild-type and cotH-null strains and analyzed spore coat extracts by means of immunoblotting with a PKC substrate-specific antibody (pPKC-Ab) that recognizes the PKC consensus phosphorylation motif. Remarkably, we observed strong immunoreactivity in proteins extracted from wild-type spores, but not from cotH-null spores (Fig. 6G). Ectopic expression of wild-type, but not the catalytically inactive mutant, in the cotH-null background increased phosphorylation of the spore coat proteins. Moreover, strong immunoreactivity was observed for proteins with estimated molecular masses of ∼66 kDa and ∼30 kDa, which we infer are CotB and CotG, respectively (Fig. 6G, asterisks). We also detected other pPKC-Ab immunoreactive bands, suggesting that CotH is likely to phosphorylate other substrates in addition to CotB and CotG. Furthermore, we cannot rule out the possibility that CotH can act as a priming kinase for an unidentified spore kinase. In any event, our results suggest that phosphorylation of coat proteins by CotH is an important regulatory mechanism for spore coat assembly and germination.
Discussion
We have discovered a family of atypical kinases that phosphorylate spore coat proteins and uncovered an unexpected function for protein phosphorylation in regulating spore coat assembly and germination. Our results suggest that phosphorylation of CotB and CotG by CotH is necessary for the proper assembly of proteins into the spore coat, which ultimately facilitates efficient germination when environmental conditions are favorable for growth. Because the spore coat structural components are synthesized in the mother cell before incorporation into the coat (38, 39), phosphorylation of CotB and CotG most likely occurs before the mature spore is developed. Indeed, previous work has demonstrated that the formation of the 66-kDa form of CotB (hyperphosphorylated) can be detected in the mother cell before a mature spore is established (36). Thus, we propose that the phosphorylation of spore coat proteins by CotH takes place before or during the incorporation of structural components into the spore coat.
Although the mechanism by which phosphorylated CotB facilitates germination is unclear, we propose that phosphorylation of CotG by CotH acts to stabilize CotG. CotG is a modular protein consisting of tandem repeats, and it may be susceptible to proteolysis in its unphosphorylated form. Therefore, loss of CotH phosphorylation would result in CotG degradation. This mechanism may explain why CotG does not accumulate in spores generated from cells depleted of CotH (26, 36). In any event, it is clear that phosphorylation is an important, but overlooked, mechanism in spore biology. Future work will be necessary to determine the mechanisms by which coat protein phosphorylation regulates germination.
The CotH kinase family is evolutionarily and structurally distinct from the His kinases of the two-component systems and only remotely related to the bacterial spore kinases (BSKs) described by Manning and coworkers (40). The BSKs belong to the choline and aminoglycoside kinase group of small-molecule kinases, some of which also reside in the spore coat, including CotS (41). The substrates for the BSKs are unknown. Furthermore, although the CotH kinases belong to the PKL fold family, they are very distant from most other PKLs, including the secreted tyrosine kinase, vertebrate lonesome kinase (VLK/PKDCC) (42), protein O-mannose kinase (POMK/Sgk196) (43), and FAM69/DIA1 family of secretory pathway kinases (44).
Analogous to the Fam20C-related kinases, many CotH proteins have signal peptides that can direct them to the extracellular space. In a survey of almost 2,000 CotH homologs, more than 30% of bacterial CotH proteins and more than 60% of eukaryotic CotH proteins contained predicted signal sequences. Thus, cell surface or extracellular roles are a common theme for the CotH kinases. Moreover, both B. subtilis CotH and Fam20C use multisite phosphorylation to modify their substrates. When these phosphorylation events are deficient, such as in B. subtilis cotH-null strain or in patients with Raine syndrome caused by Fam20C mutations, the result is impaired assembly of extracellular macromolecular structures, such as the spore coat or mineralized tissues. Therefore, understanding how phosphorylation of spore coat proteins by CotH contributes to the assembly and germination of spores is likely to shed light on analogous fundamental mechanisms that eukaryotic cells use to build extracellular structures, such as bone and teeth.
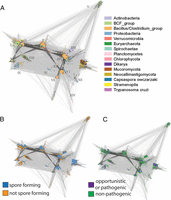
CotH homologs are present in many bacterial and eukaryotic species, with the broadest diversity observed in the bacterial Bacillus/Clostridium group, followed by fungi (Fig. 7A). One well-separated cluster comprises only fungal Mucoromycota and Neocallimastigomycota sequences, but not those from higher fungi. A few fungal sequences from Dikarya, together with multiple eukaryotic Phytophtora, Chlamydomonas, and Capsaspora sequences, are located in a large cluster, together with many bacterial sequences from distant lineages. It is noteworthy that the eukaryotic CotH sequences are not clustered together. Rather, they form lineage-specific subgroupings with bacterial sequences, pointing at possible horizontal gene transfer (HGT). The overall pattern seems to be a consequence of multiple gene duplications at various time points, with subsequent gene loss and HGT between distant organisms. Furthermore, sporulation and pathogenicity are common, but not obligatory, in CotH-possessing organisms (Fig. 7 B and C). Indeed, a role in host cell invasion in mucormycosis was shown for a fungal CotH protein (45), and although pathogens such as B. anthracis are notable among CotH-containing organisms, many others are nonpathogenic.
Taxonomy, sporulation, and pathogenicity of CotH-possessing organisms. (A) CLANS clustering analysis of CotH homologs represented graphically by the network of BLAST-derived sequence similarities (edges) between representative CotH proteins (dots). Clusters are colored by taxonomy. Roman numerals denote sequence clusters (representatives in the alignment are shown in Fig. S2). CLANS clustering analysis of CotH homologs is colored by ability to sporulate (B) or by pathogenicity (C).
The distant sequence relationships between CotH and other PKL proteins suggest early divergence of the FAM20/HipA/CotH family group from the main “core” of the PKL clan (Fig. S1). The dominance of bacterial taxa among CotH-possessing organisms suggests a bacterial origin for CotHs with a single transfer to Archaea and multiple transfers to Eukaryotes. However, an ancestral eukaryotic CotH cannot be ruled out, with at least four distinct eukaryotic lineages carrying cotH copies: green algae, fungi, stramenopiles, and trypanosomes. This picture is further complicated by possible HGT events involving cotH genes.
A generic hypothesis regarding the functional roles of uncharacterized CotH proteins may be formed using the presence of additional protein domains present in some CotHs. The CotH kinase-like domains are often preceded and/or followed by domains consisting predominantly of beta-sheet, such as fibronectin-like (Pfam: PF13287), lamin tail (Pfam: PF00932), chitobiase/beta-hexosaminidase C-terminal (Pfam: PF03174), or cellulose-binding domains (Pfam: PF00553). Also, dockerin type I repeats (Pfam: PF00404) are found in some CotHs, which are involved in the formation of the cellulosome. Most of these domains are typically involved in carbohydrate binding. In some cases, CotH kinase domains coexist with catalytic domains that act on carbohydrates, pectate lyases, or glycosyl hydrolases. Accordingly, a generic role in cell–wall interaction and manipulation can be postulated for some CotHs.
In summary, we have discovered a family of protein kinases distantly related to the Fam20C-family of secretory pathway kinases and uncovered a previously unidentified role for protein phosphorylation in spore biology. Because several CotH-containing organisms are human pathogens, our work may also have important clinical implications to combat human diseases such as anthrax and mucormycosis.
Materials and Methods
Bioinformatics.
For elucidation of distant sequence similarity between CotH proteins and PKL families, three methods were used, namely, fold and function assignment system 03 (FFAS03) (46), which uses sequence profile-to-profile comparison; homology detection and structure prediction (HHpred) (47), which employs hidden Markov model (HMM)-to-HMM comparison; and Phyre (48), which is a metapredictor exploiting several independent algorithms. Standard parameters and significance thresholds were used. For visual clustering of families within the broad PKL clan, the cluster analysis of sequences (CLANS) algorithm (49) was performed on a set of sequences, including (i) all of the Pfam seeds from the 19 families of the PKL clan (Pfam identifier CL0016) and (ii) other proteins with known or predicted structural similarity to the PKL clan. This included all of the Pfam seeds from the Pfam families UPF0061/SELO, PIP49_C/FAM69, Alpha_kinase, PI3_PI4_kinase, Act-Frag_cataly, PPDK_N, PIP5K, HipA, DUF1193/FAM20, and FAM198. CLANS was run with three iterations of position-specific iterated (PSI)-BLAST, using the blocks substitution matrix 45 (BLOSUM45) substitution matrix and inclusion threshold of 0.001. For the graph, similarity relations with P value significance below 0.1 were considered. Some alignments of kinase-like families in Pfam did not include complete kinase domains and had to be corrected and extended manually (e.g., UL97, PIP49_C, HipA).
Signal peptides were predicted by the SignalP server (50). Multiple sequence alignments of the CotH domains were constructed by using the PROMALS (51) and MUSCLE programs (52) and visualized with Jalview (53). Phylogenetic analysis of CotH and representatives of selected kinase families was performed using the phylogeny.fr server (54), using the maximum likelihood method PhyML, with the approximate likelihood-ratio test for branch support estimation, and starting from a PROMALS multiple sequence alignment. Sequence variability was displayed as sequence logos using the WebLogo server (55). For Pfam domain assignments, HMMER3 (56) on the Pfam (57) database was used.
For the survey of diversity within the CotH family, searches were initiated using the B. anthracis CotH protein sequence. Sequences from jackhmmer (58) searches [executed using the standard parameters on the nonredundant (NR) databases as of May 2014] and web-based BLAST searches against Joint Genome Institute fungal genomes database (59) generated the CotH sequence dataset, containing 1,735 sequences. This dataset was manually annotated with lifestyle features related to pathogenicity and spore/resting form/capsule formation. CotH domain sequences were clustered with CLANS (49). Pairwise BLAST search results were used when the P value significance was greater than 0.001. The same clustering result was alternatively colored according to taxonomical information (National Center for Biotechnology Information order/phylum), pathogenic/nonpathogenic lifestyle, and the ability to form spores.
Reagents.
MyeBP and H2A were from Sigma. Anti-V5 antibody was from Invitrogen (catalog no. 46-0705). Anti-phospho PKC substrate antibody was from Cell Signaling (catalog no. 2261S). Masaya Fujita (University of Houston, Houston, TX) generously provided the anti-SigA antibody. Recombinant OPN was prepared as previously described (5). Selenomethionine medium was purchased from Molecular Dimensions (catalog no. MD12-500).
Generation of Constructs.
The B. subtilis CotH and CotB coding sequences were amplified by PCR using B. subtilis (strain PY79) genomic DNA as a template. The B. cereus CotH coding sequence was synthesized as a gBlock (60) and used as a template for PCR. The amplified ORFs were cloned into a modified pET28a bacterial expression vector (ppSumo), containing an N-terminal 6× His tag, followed by the yeast Sumo (smt3) coding sequence. The CotB coding sequence was cloned in the pet28a vector.
Protein Purification.
E. coli BL21 (DE3) competent cells were transformed with ppSumo-B. subtilis CotH, B. cereus CotH, or pET21a-CotB, and the cells were grown in Luria–Bertani medium at 37 °C until the A600 reached ∼0.5–0.7. Protein expression was induced by addition of 0.4 mM isopropyl β-d-thiogalactopyranoside overnight at 30 °C. The cells were recovered by centrifugation at 5,000 × g for 10 min, resuspended in lysis buffer [50 mM Tris⋅HCl (pH 7.5–8.0), 250–300 mM NaCl, 1 mM PMSF, 1× protease inhibitor mixture, and 0.1% β-mercaptoethanol], and lysed using sonication. For B. subtilis and B. cereus CotH, the cell lysates were centrifuged at 35,000 × g for 45 min and the 6× His-tagged fusion proteins were purified from the supernatant by Ni2+-NTA-agarose chromatography. The resin was washed extensively with lysis buffer containing 25 mM imidazole, and the fusion proteins were eluted with lysis buffer containing 300 mM imidazole. The proteins were cleaved overnight at 4 °C with 3 μg/mL recombinant 6×-His tagged Ulp1 Sumo protease. The proteins were further purified by gel filtration chromatography using an AKTA Pure FPLC chromatography system (GE Healthcare).
For B. subtilis CotB, 1% SDS was added to the lysis buffer to aid in solubilization of the protein (61). The cells were lysed by sonication, followed by chilling to precipitate the SDS. The lysate was centrifuged and purified as above.
In Vitro Kinase Assays.
In vitro kinase assays were typically performed in a reaction mixture containing 50 mM Tris⋅HCl (pH 7.5); 5 mM MnCl2; 0.5 mM [γ-32P]ATP (specific activity = 100–500 cpm/pmol); 0.25 mg/mL MyeBP, H2A, or OPN; and 10 μg/mL B. subtilis or B. cereus CotH. Reactions were incubated at 30 °C and terminated at the indicated time points by the addition of EDTA, SDS loading buffer, and boiling. Reaction products were separated by SDS/PAGE and visualized by Coomassie blue staining. Destained gels were dried, and incorporated radioactivity was visualized by autoradiography.
The peptide HKKSYRSHKKSRS, CotG(88–100) was synthesized by standard O-(Benzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate/1-hydroxybenzotriazole Fmoc solid-phase chemistry as described (12). Kinase assays were performed in 50 mM Hepes-KOH (pH 7); 5 mM MnCl2, MgCl2, CoCl2, CaCl2, FeCl2, or ZnCl2; 1 mM CotG(88–100); 0.5 mg/mL BSA; 500 μM [γ-32P]ATP (specific activity = 100–500 cpm/pmol); and 10 μg/mL B. subtilis CotH wild-type or mutants for 10 min at 30 °C. For the experiments performed in Fig. 4D, Sumo-tagged CotH wild-type or mutants were used in the assays with 5 mM MnCl2 at a concentration of 25 μg/mL. Preliminary experiments were performed using these conditions to ensure the reactions were linear with respect to time. Reaction mixtures were spotted on P81 phosphocellulose filters and terminated by immersion in 75 mM H3PO4. Filters were washed three times for 15 min each time, followed by immersion in acetone for 5 min. After air-drying, incorporated radioactivity was quantified by Cerenkov counting in a Beckman LS 6000IC scintillation counter. For time courses, the reactions were first stopped by the addition of 20 mM EDTA and the samples were analyzed as above.
Crystallization and X-Ray Data Collection.
Recombinant B. subtilis CotH in 25 mM Tris⋅HCl (pH 7.5) and 50 mM NaCl was concentrated to 5 mg/mL. The crystals were grown at 20 °C by the sitting drop vapor diffusion method using a 1:1 ratio of protein/reservoir solution containing 13% (wt/vol) PEG3350, 0.1 M cacodylate (pH 6.75), and 0.2 M MgCl2. Crystals were flash-frozen in 15% (wt/vol) PEG3350, 0.1 M MES (pH 6.75), 0.2 M MgCl2, 50 mM NaCl, and 30% (vol/vol) ethylene glycol. Selenomethionine (SeMet)-labeled protein was obtained by expressing B. subtilis CotH in B834 cells grown in SeMet media. AMP-bound CotH was obtained by incubating protein with 10 mM ADP and 2 mM AIF3. The crystals were grown at 20 °C by the sitting drop vapor diffusion method using a 1:1 ratio of protein/reservoir solution containing 12% (wt/vol) PEG3350, 0.1 M cacodylate (pH 6.75), and 0.2 M MgCl2. Crystals were flash-frozen in 15% (wt/vol) PEG3350, 0.1 M MES (pH 6.75), 0.2 M MgCl2, and 30% (vol/vol) ethylene glycol.
Diffraction data were collected at Advanced Photon Source beamline 19-ID using incident radiation at the Se K-edge. All data were collected at 100 K. Data were indexed, integrated, and scaled using the HKL-3000 program package (62). Native and selenomethionyl-derivatized CotH crystals exhibited the symmetry of space group P212121, with unit cell parameters of a = 53 Å, b = 63 Å, and c = 118 Å, and contained one molecule of CotH per asymmetrical unit. Data collection statistics are provided in Table S1.
Phase Determination and Structure Refinement.
Phases were obtained from a single wavelength anomalous dispersion experiment using selenomethionyl-derivatized CotH with data to a minimal d-spacing of 1.47 Å. Nine selenium sites were located, phases were improved, and an initial model containing 96% of all CotH residues was automatically generated in the AutoBuild routine of the Phenix program suite (63).
Because the selenomethionyl-derivatized and native crystals were isomorphous, all further calculations were performed vs. the native data. Additional residues for CotH were manually modeled in the program Coot (64). Positional and isotropic atomic displacement parameter refinement, as well as translation, libration, skew (TLS) atomic displacement parameter refinement, was performed to a resolution of 1.63 Å for the apo-bound CotH and to a resolution of 1.40 Å for the Mg2+/AMP-bound CotH using the program Phenix with a random 3% of all data set aside for an Rfree calculation. Phasing and model refinement statistics are provided in Table S1.
Immunoblotting.
Mother cell and spore extract proteins were separated by SDS/PAGE, transferred to nitrocellulose membranes, stained with Ponceau S, and then blocked in 5% (wt/vol) milk or 4% (wt/vol) BSA. The membranes were probed with anti-V5 (1:1,000), anti-SigA (1:5,000), or anti-phosphoPKC substrate (1:1,000) antibodies for 1–2 h at room temperature or overnight at 4 °C. Detection was performed by enhanced chemiluminescence, followed by autoradiography.
SI Materials and Methods
Generation of Bacillus subtilis Strains.
Bacillus subtilis strain PY79 was used to generate all of the strains in this study. The strains and primers used in this study are listed in Tables S2 and S3. Strains were generated by transformation of competent B. subtilis cells with plasmid or genomic DNA as previously described (66). Strains were verified by antibiotic selection, PCR, and sequencing of PCR amplification products. More details on plasmid construction are presented in Plasmid Construction, below.
Germination Assays.
Spore germination was measured by loss of OD580 over time (67). Spores were heat-activated in water, pelleted, and resuspended in 10 mM Tris⋅HCl (pH 8.4), 1 mM d-glucose, 1 mM d-fructose, and 10 mM KCl to a final OD580 of ∼0.3. Spores were equilibrated at 37 °C before induction of germination by the addition of 10 mM L-alanine. OD580 readings were taken at 5-min intervals until a constant reading was achieved.
Sporulation Time Point and Extraction of Mother Cell Proteins.
Sporulation was induced by the resuspension method (68), but strains were grown in 25% (vol/vol) Luria–Bertani medium before resuspension (69). Samples were harvested at different time points (70), pelleted, resuspended in a modified buffer with 25 mM Tris⋅HCl (pH 7.5), 100 mM NaCl, 1 mM EDTA, 15% (vol/vol) glycerol, 1 mM PMSF, 1 mg/mL lysozyme, and 1× protease inhibitor mixture. Following incubation on ice for 20 min, samples were disrupted by sonication (four 20-s pulses). The supernatant was collected by centrifugation (12,000 × g for 10 min at 4 °C) and boiled in SDS loading buffer before separation by SDS/PAGE.
Mass Spectrometry.
The CotG(88–100) peptide (1 mM) was incubated with 10 μg/mL B. subtilis CotH overnight at 30 °C. The reaction was terminated by the addition of 18.5 mM EDTA. Peptides were analyzed by high-pressure liquid chromatography coupled with tandem mass spectroscopy (LC-MS/MS) by the University of California, San Diego Biomolecular and Proteomics Mass Spectrometry Facility using nanospray ionization as previously described (71) with the following changes: The nanospray ionization experiments were performed using a TripleTof 5600 hybrid mass spectrometer (ABSCIEX) interfaced with nanoscale reversed-phase ultraperformance liquid chromatography (nanoACQUITY; Waters Corporation) using a 20-cm × 75-μm i.d. glass capillary packed with 2.5-μm C18 (130) CSHTM beads (Waters Corporation). Peptides were eluted from the C18 column into the mass spectrometer using a linear gradient [5–80% (vol/vol)] of acetonitrile (ACN) at a flow rate of 250 μL/min for 1 h. The buffers used to create the ACN gradient were as follows: buffer A [98% (vol/vol) H2O, 2% (vol/vol) ACN, 0.1% formic acid, and 0.005% TFA] and buffer B [100% (vol/vol) ACN, 0.1% formic acid, and 0.005% TFA]. MS/MS data were acquired in a data-dependent manner in which the MS1 data were acquired for 250 ms at an m/z of 400–1,250 Da and the MS/MS data were acquired from an m/z of 50–2,000 Da. The independent data acquisition parameters were as follows: MS1-TOF acquisition time of 250 ms, followed by 50 MS2 events of 48 ms acquisition time for each event. The threshold to trigger the MS2 event was set to 150 counts for ions with a charge state of +2, +3, and +4. The ion exclusion time was set to 4 s. Finally, the collected data were analyzed using Protein Pilot 4.5 (ABSCIEX) for peptide identifications.
Plasmid Construction.
pcotH::kanr.
This plasmid was constructed by Gibson assembly using the following four DNA fragments: (i) a DNA fragment of 765 bp amplified with primers JLG-1000 and JLG-1001 from genomic DNA of B. subtilis PY79, encompassing the 5′ region of the cotG coding sequence through the 5′ region of the cotH coding sequence; (ii) a fragment generated with primers JLG-7 and JLG-97 from pJLG3 that contains a kanamycin-resistant gene (72); (iii) a fragment of 888 bp amplified with primers JLG-1007 and JLG-1008 from genomic DNA of B. subtilis PY79, encompassing the 3′ region of the cotH coding sequence through the 5′ region of the cotB coding sequence; and (iv) a fragment amplified with primers JLG-95 and JLG-96, encompassing the spectinomycin-resistant gene, the origin of replication, and the ampicillin-resistant gene from pDG1662 (73).
pJLG13-cotHB.
This plasmid was constructed by Gibson assembly using the following two DNA fragments: (i) a DNA fragment of 2,863 bp amplified by primers JLG-998 and JLG-1006 from genomic DNA of B. subtilis PY79, encompassing the 5′ region of the cotG coding sequence and the entire coding sequences of cotH and cotB, and (ii) a fragment amplified by inverse PCR of pJLG13 (72) with primers JLG-139 and JLG-215, encompassing an ampicillin-resistant gene, a spectinomycin-resistant gene, the origin of replication, and a chloramphenicol-resistant gene.
pJLG13-cotHD251A-cotB.
This plasmid was constructed using Quickchange mutagenesis of pJLG13-cotHB with primers VST-3 and VST-4.
pJLG13-cotGHB/pJLG13-cotG-HD251A-cotB.
These two plasmids were constructed by Gibson assembly using the following three DNA fragments: (i) a DNA fragment of 3,482 bp amplified with primers KBN-30 and KBN-27 of pJLG-cotHB; (ii) a fragment of 6,339 bp amplified with primers KBN-26 and KBN-31, encompassing the 5′ coding sequence of cotG and the entire coding sequences of cotH and cotB of pJLG-cotHB or pJLG13-cotHD251A-cotB; and (iii) a fragment of 769 bp amplified with primers KBN-28 and KBN-29 from genomic DNA of B. subtilis PY79, encompassing the 5′ coding sequence of ywrF through the 3′ coding sequence of cotG.
pJLG13-PcotG-PcotH-cotH.
This plasmid was constructed by Gibson assembly using the following two DNA fragments: (i) a DNA fragment of 1,334 bp amplified with primers JLG-1006 and JLG-1087 from genomic DNA of B. subtilis PY79, encompassing cotH coding sequence and putative promoter immediately upstream of cotH (25), and (ii) a fragment generated by inverse PCR of pJLG13-cotGHB with primers JLG-139 and JLG-1086. This construction places cotH under the control of its native promoters: PcotG (74) and an additional putative promoter immediately upstream of cotH coding sequence (25).
pJLG13-PcotG-PcotH-cotH-V5/pJLG13-PcotG-PcotH-cotHD251A-V5.
These two plasmids were constructed by Gibson assembly using the following three DNA fragments: (i) a DNA fragment of 1,250 bp with primers KBN-164 and KBN-172 from pJLG13-cotHB or pJLG13-cotHD251A-cotB, containing part of the cotH coding sequence; (ii) a 3,497-bp fragment with primers KBN-166 and KBN-167 from pJLG13-PcotG-PcotH-cotH; and (iii) a 3,975-bp fragment with primers KBN-173 and KBN-169 from pJLG13-PcotG-PcotH-cotH.
pamyE::PgerE-cotHD251A-V5.
This plasmid was constructed by Gibson assembly using the following two DNA fragments: (i) a DNA fragment corresponding to the 631 bp immediately upstream of gerE start codon, amplified with primers JLG-1241 and JLG-1242 from genomic DNA of B. subtilis PY79, and (ii) a fragment generated by inverse PCR of pJLG13-PcotG-PcotH-cotHD251A-V5 with primers JLG-215 and JLG-1240.
pamyE::PcotE-cotHD251A-V5.
This plasmid was constructed by Gibson assembly using the following two DNA fragments: (i) a DNA fragment corresponding to the 107 bp immediately upstream of cotE start codon, amplified with primers JLG-1246 and JLG-1247 from genomic DNA of B. subtilis PY79, which encompasses the σK-dependent promoter of cotE (P2), but not the σE-dependent promoter (P1) (75), and (ii) a fragment generated by inverse PCR of pJLG13-PcotG-PcotH-cotHD251A-V5 with primers JLG-215 and JLG-1240.
pamyE::PcotA-cotHD251A-V5.
This plasmid was constructed by Gibson assembly using the following two DNA fragments: (i) a DNA fragment corresponding to the 417 bp immediately upstream of cotA transcription start site, amplified with primers JLG-1243 and JLG-1244 from genomic DNA of B. subtilis PY79, and (ii) a fragment generated by inverse PCR of pJLG13-PcotG-PcotH-cotHD251A-V5 with primers JLG-215 and JLG-1245.
Spore Purification and Spore Coat Extraction.
Sporulation was induced by the exhaustion method in Difco sporulation medium (67). Strains were grown for 48–72 h and then collected and washed with 4 °C sterile water; the spores were incubated in sterile water at 4 °C to lyse any remaining vegetative cells. Spores were further purified over a phosphate-polyethylene glycol aqueous biphasic gradient (76). The top organic phase containing the spores was removed and extensively washed in at least 50 vol of 4 °C sterile water. The spores were pelleted, resuspended in sterile water, and stored at 4 °C. Sample purity was checked using phase contrast microscopy. Spore samples of >95% purity (phase-bright spores) were used in the studies.
Spore coat extracts were prepared as previously described using a modified detergent buffer (67). Spore numbers were normalized at OD580, pelleted, resuspended in 1.5% (wt/vol) SDS and 50 mM DTT, and then incubated at 70 °C for 30 min. The supernatant was collected by centrifugation (10,000 × g for 10 min at room temperature) and boiled in SDS loading buffer before separation by SDS/PAGE.
Acknowledgments
We thank Greg Taylor, Kim Orth, and members of the V.S.T. and J.E.D. laboratories for insightful discussion; David King for generously synthesizing peptides for this work; Lisa Kinch, Junyu Xiao, and Michael Reese for valuable input on the structure; and Jose Cabrera for help with the figures. Results shown in this report are derived from work performed at the Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. The Argonne National Laboratory is operated by the University of Chicago Argonne, LLC, for the US Department of Energy, Office of Biological and Environmental Research, under contract DE-AC02-06CH11357. This work was supported by NIH Grants DK018849 and DK018024 (to J.E.D), R00DK099254 (to V.S.T.), and GM57045 (to K. Pogliano), and the Howard Hughes Medical Institute. K. Pawlowski was supported by a grant from the Polish National Science Centre (2012/05/B/NZ3/00413). A.M. was supported by a grant from the National Science Centre scholarship for outstanding young researchers, Ministry of Science and Higher Education (2012/07/D/NZ2/04286). V.S.T. is the Michael L. Rosenberg Scholar in Medical Research and a Cancer Prevention Research Institute of Texas Scholar (RR150033).
Footnotes
-
↵1K.B.N. and A.S. contributed equally to this work.
- ↵2To whom correspondence may be addressed. Email: jedixon{at}ucsd.edu or vincent.tagliabracci{at}utsouthwestern.edu.
-
Author contributions: K.B.N., A.S., E.S.D., J.L.-G., A.M., M.D., S.Y., D.R.T., K. Pawlowski, and V.S.T. designed research; K.B.N., A.S., E.S.D., J.L.-G., A.M., M.D., M.G., S.Y., D.R.T., K. Pawlowski, and V.S.T. performed research; K.B.N., A.S., E.S.D., J.L.-G., A.M., M.D., M.G., S.Y., K. Pogliano, D.R.T., K. Pawlowski, J.E.D., and V.S.T. analyzed data; and K.B.N., A.S., J.E.D., and V.S.T. wrote the paper.
-
Reviewers: J.S., University of Washington School of Medicine; and A.S., Genentech.
-
The authors declare no conflict of interest.
-
Data deposition: The structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 5JD9 and 5JDA).
-
See Commentary on page 6811.
-
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1605917113/-/DCSupplemental.